Multiomics analyses
This is an integrative approach to combine data sets of different omic groups during analysis. Multiomic studies apply a range of techniques—including genomics, transcriptomics, proteomics and metabolomics to generate a comprehensive understanding of the biological process in question. A typical multiomics workflow combines transcriptomics and proteomics from varying experimental conditions to address biological questions, followed by validation experiments to verify or nullify conceived hypotheses.
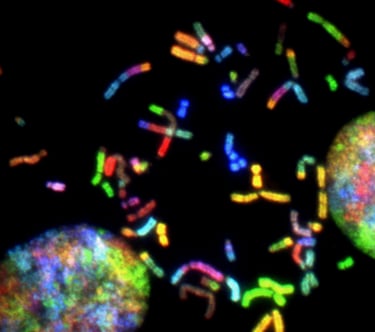
RNA-seq sample

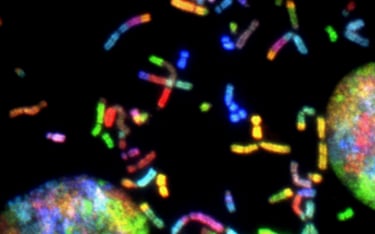
Proteomics sample
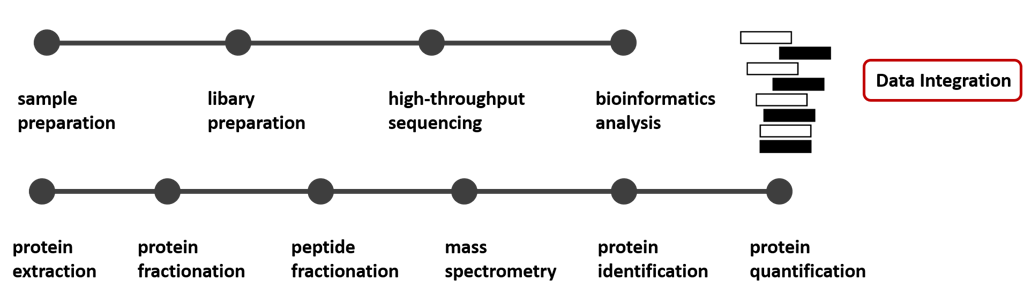
Typical multiomics workflow
Metabolomics
Metabolomics is the comprehensive analysis of small molecules (metabolites) that constitute the downstream products of cellular regulatory processes. These metabolites include intermediates of central carbon metabolism, amino acids, nucleotides, cofactors, and lipids, thereby reflecting the integrated outcome of gene expression, protein activity, and environmental influences. As such, metabolomics provides a direct functional readout of biological state under defined conditions.
In microbiology, metabolomics enables the systematic investigation of metabolic regulation, stress adaptation, antimicrobial resistance, host–pathogen interactions, and virulence. By capturing rapid biochemical changes that may not be evident at the transcriptomic or proteomic level, metabolomics provides essential functional context for interpreting molecular mechanisms. Multiomics integration with transcriptomics and proteomics supports systems-level analysis of pathway activity, regulatory networks, and metabolic reprogramming.
Furthermore, lipidomics represents a specialized and integral component of metabolomics, focusing on the detailed characterization of lipid species involved in membrane organization, energy storage, and cellular signalling. Given the structural diversity and biological relevance of lipids, lipid-focused analyses enable high-resolution insight into membrane dynamics, metabolic remodelling, immune modulation, and pathogen adaptation.
Untargeted metabolomics is a hypothesis-generating approach aimed at global profiling of metabolites without prior selection of analytes. This strategy enables the detection and relative quantification of a wide range of metabolites across diverse chemical classes, including lipids. Untargeted analyses are particularly valuable for exploratory studies, comparative phenotyping, and the identification of unexpected metabolic alterations, novel biomarkers, and previously unrecognized pathway perturbations.
Targeted metabolomics is a hypothesis-driven approach focused on the accurate and reproducible quantification of predefined metabolites or metabolic pathways with established biological relevance. Using optimized analytical workflows and internal standards, targeted strategies provide high sensitivity, specificity, and quantitative robustness. This approach is well suited for pathway validation, mechanistic investigations, and comparative analyses across defined biological conditions, such as microbial adaptation, infection models, and treatment responses.
Sample types for multiomics




cell lines
tissue
lysates, biological fluids
extracellular vesicles
microbial cultures


plant materials


